CRISPR/Cas9 complications
This subpage constitutes the fifth part of the theory for Biotech Academy’s material on CRISPR-Cas9.
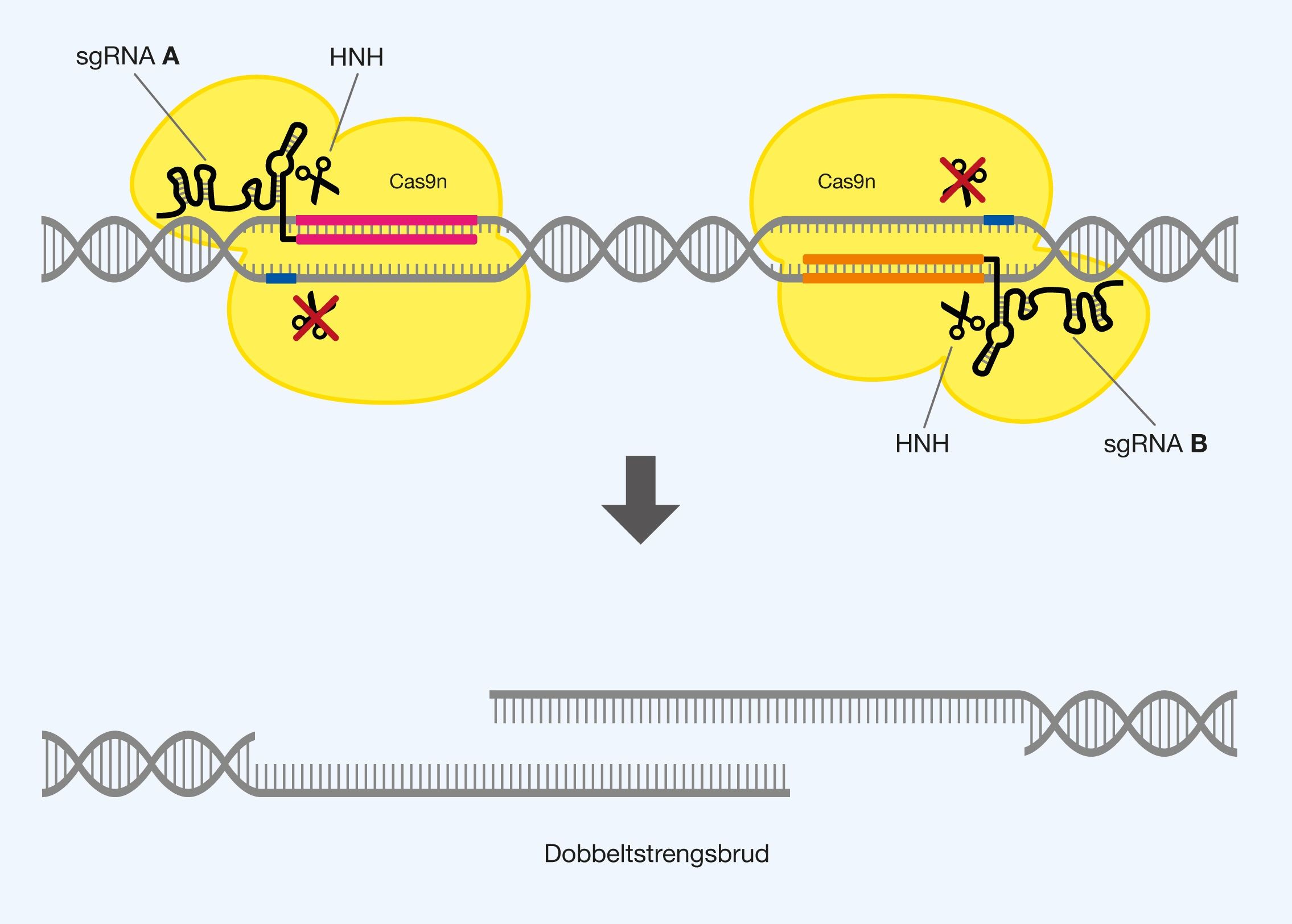
Off-target effects and possible solutions
The first problem encountered with genetic modification with Cas9 is off-target effects. These are seen when the Cas9 protein interacts with a different sequence than the one planned to modify. It is clear that this can create major problems, as, for example, important genes can be destroyed unconsciously. The problem arises mainly because the sequence that the given sgRNA recognizes exists in several places in the genome, or that there are sequences that are sufficiently similar to it. The alternate PAM sequences 5′-NAG-‘3 and 5′-NGA-3’ may also be recognized to a lesser extent, which lowers specificity. Off-target effects are difficult to predict for individual sequences in a genome, which increases uncertainty when using Cas9. You can reduce the amount of off-target effects by trying to make sure that your sgRNA is as unique as possible in relation to one sequence in the genome. With the specificity from the PAM sequence and the 20 recognizing nucleotides, it is easy to achieve in organisms with small genomes, but can be much more complicated in complex organisms with large genomes, such as humans.